US Pharm. 2009;34(9):HS9-HS13.
There are over 350,000 cases of culture-positive nosocomial bloodstream infections (BSIs) in the United States each year, costing approximately $27,000 per community-acquired BSI and between $58,000 and $101,000 per hospital-acquired BSI.1-4 Early treatment of BSIs with appropriate antimicrobial therapy has proven to reduce morbidity, mortality, and health care costs.1,5 Current microbiological techniques used for pathogen identification and susceptibility are phenotypic (i.e., observable trait) methods, which require time for bacterial growth. Due to this inherent limitation, genotypic (i.e., genetic makeup) methods of identifying pathogens have been introduced from the laboratory into clinical practice, promising faster results and improved patient outcomes and subsequent decreased health care costs. While some hospitals have acquired various rapid molecular testing techniques to quickly identify BSIs, few published studies have evaluated the clinical impact and cost-effectiveness of implementing these tests in the clinic setting.
Types of Rapid Molecular Testing
There are currently several general types of techniques for rapidly identifying pathogens associated with BSIs on a genotypic basis (TABLE 1). All of these techniques detect pathogens by testing for the presence of genus- and/or species-specific expressed gene products. However, the way in which each one does this is different.
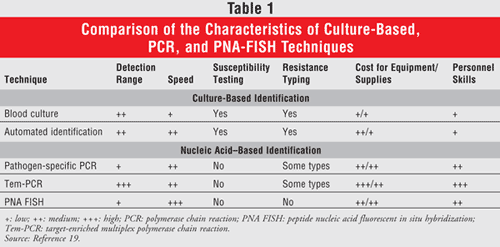
Polymerase Chain Reaction (PCR) Techniques: Amplification techniques work by amplifying a small, specific sequence of DNA, then measuring the amount of fluorescent signal that is created when specific fluorescence resonance emission transfer probes, molecular beacons, or TaqMan (hydrolysis) probes bind to the amplified gene product being tested. Many of these gene products can be identified within 2 hours. Real-time polymerase chain reaction (RT-PCR) has been increasingly utilized in the clinical setting for a number of different applications, including detecting the presence of Clostridium difficile toxins from feces, quickly identifying slowly cultivable organisms, and detecting the presence of genes associated with antimicrobial resistance.6 Because it is only capable of testing for one specific gene product per sample, the test can only be cost-effective after the microbiology laboratory has identified the genus and whether the pathogen is a gram-positive or gram-negative organism. Although being able to identify Staphylococcus aureus, rule out coagulase-negative staphylococcal species, and determine if S aureus is methicillin resistant (MRSA) or sensitive (MSSA) can be accomplished faster using RT-PCR, this method still relies on time-consuming standard culture techniques to narrow down which organisms are in question. RT-PCR is Clinical Laboratory Improvement Amendments (CLIA) certified and FDA approved, making it eligible for reimbursement from the Centers for Medicare and Medicaid Services (CMS) and ensuring quality laboratory testing. Whether or not this narrow application is cost-effective is not yet known.
Advancements in PCR technology have overcome the limitations of RT-PCR with the development of target-enriched multiplex polymerase chain reaction (Tem-PCR) and other similar multiplex PCR techniques, which allow for the detection of multiple specific gene products from a single sample in less than 5 to 6 hours.7,8 This gives Tem-PCR and other multiplex PCR techniques the opportunity to test for the presence of common viruses, pathogenic bacteria, yeast, and any known genes associated with antimicrobial resistance from a single sample, negating the need for the microbiology lab to culture the sample to determine the specific genes. The major drawbacks of Tem-PCR technology are that it is costly, requires trained technicians, and may require samples to be tested off-site rather than in the hospital microbiology laboratory because this test is CLIA certified, yet not FDA approved.
PNA FISH: Another recent type of rapid testing technique is peptide nucleic acid fluorescent in situ hybridization (PNA FISH), which is both CLIA certified and FDA approved. It is similar to nucleic acid amplification techniques in that it uses fluorescein-labeled probes, but it differs by binding to the 16S rRNA gene of the live bacteria rather than binding to amplified gene products.9 Once bound, a fluorescent signal is created and the sample is read under a fluorescent microscope to determine which species are present, based on morphological characteristics of the live bacteria and the specific fluorescent color associated with each species.9 Clinical studies utilizing PNA FISH to identify Staphylococcus and Candida species have noted a sensitivity and specificity of 100%, making this a reliable technique for use in the clinical setting.10,11 This is considered the simplest type of rapid microbiological testing and does not provide any information about the presence or absence of antimicrobial resistance genes. Another drawback to PNA FISH testing, similar to that of RT-PCR, is that it can only differentiate one genus at a time, giving it a narrow application and requiring standard culture techniques to direct which test to use in order to be cost-effective. Additionally, a specific type of microscope is needed in the hospital laboratory to determine the fluorescent results produced by this testing technique. PNA FISH currently offers FDA-cleared tests for the identification of S aureus, coagulase-negative Staphylococcus species, Enterococcus faecalis, Candida albicans, Candida glabrata, Escherichia coli, and Pseudomonas species (as well as several groups of organisms E coli/Klebsiella and C albicans/tropicalis) directly from positive blood cultures.
Potential Cost Savings
Despite the increasing number of rapid molecular and microbiological testing techniques being implemented in hospital microbiology laboratories, there are a limited number of published studies justifying their cost-effectiveness and clinical utility. Previous studies have noted a significant decrease in overall hospital costs per patient when BSIs are initially treated appropriately, compared to those not receiving initial appropriate treatment. While many rapid molecular and microbiological techniques describe a potential cost savings based on faster results, PNA FISH currently has the most published data justifying its use on a clinical and economic basis.10-12
A retrospective, cost-effectiveness analysis done by Forrest et al in 2006 evaluated the impact of rapid differentiation of S aureus from coagulase-negative staphylococci (CoNS) with PNA FISH in blood culture samples positive for gram-positive cocci in clusters (GPCC).10 The study found a statistically significant difference in patients in the PNA FISH group who received one or fewer doses of vancomycin (43% vs. 15%; P ≤.005), a decrease in median length of stay (LOS) (4 days vs. 6 days; P ≤.05), and an overall cost savings of approximately $4,000 per patient ($9,616 vs. $13,621).
When Forrest et al evaluated the impact of PNA FISH rapid identification of C albicans on the selection and expenditure of antifungal therapy, the study found that this test significantly reduced the median time required for identification compared to standard techniques by 34.5 hours (9.5 hours vs. 44 hours; P ≤.001) and reduced the defined daily dosage (DDD) per patient usage of caspofungin (3.2 vs. 8.7; P <.05), with a resulting cost savings of $1,978 per patient.11 After accounting for the cost of PNA FISH testing materials, the overall cost savings was $1,729 per patient.
Ly et al evaluated further decreasing the time to reporting PNA FISH data in patients with blood culture samples positive for GPCC by adding a clinical laboratory liaison who rapidly relayed the information to physicians within 3 hours of the results.12 Use of the laboratory clinical liaison reduced overall mortality (16.8% control group vs. 7.9% active group; P = .05) with the biggest impact seen in intensive care unit (ICU) patients (47.8% control group vs. 9.5% active group; P = .01). Use of a laboratory clinical liaison also reduced further overall antibiotic use by 2 days (P = .01) and antimicrobial use in patients with blood cultures positive for CoNS by 2.5 days (P = .01). Early notification of PNA FISH results reduced the LOS in patients with CoNS by 2 days in the non-ICU setting and 7 days in the ICU and subsequently showed a trend towards reduced median hospital charges of $19,441 per patient (P = .09).
A quasiexperimental study done by Forrest et al assessed the utility of PNA FISH in patients with enterococcal BSIs.13 The primary objective was to determine if PNA FISH would lead to earlier initiation of appropriate antimicrobial therapy for patients with Enterococcus faecium or E faecalis bacteremia. Secondary objectives included mortality and length of stay.10 Patients were derived from two consecutive time periods. In 2005, E faecium and E faecalis were identified using standard microbiological methods, and in 2006 they were identified using PNA FISH and standard microbiological methods. An antimicrobial management team (AMT), consisting of an infectious disease pharmacist and a physician, followed patients in both groups and used a treatment algorithm based on their institution’s antibiogram. The AMT directed therapy based on clinical factors and final sensitivity reports in the preintervention group, and intervened at the time of PNA FISH results to direct antimicrobial therapy in the intervention group. A total of 224 patients were included, with 129 in the preintervention group and 95 in the intervention group. PNA FISH identified E faecalis 2.9 days earlier than conventional culture methods (1.1 versus 4 days; P <.001); however, most patients received effective empirical therapy in both the preintervention group and intervention group (99% vs. 96%, respectively; P = .4), and there was no difference in mortality between the groups (13% vs. 10%; P = .73). E faecium was identified an average of 2.3 days earlier using PNA FISH (1.1 vs. 3.4 days; P <.001) and was associated with statistically significant reductions in the time to initiating effective therapy (1.3 vs. 3.1 days; P <.001) and decreased 30-day mortality (26% vs. 45%; P = .04). Cost savings associated with using PNA FISH in this population were not reported. The authors concluded that the use of PNA FISH in conjunction with an AMT and treatment algorithm led to earlier identification of Enterococcus species and earlier initiation of effective treatment in patients with hospital-acquired enterococcal bacteremia.13
Improving the Value of Clinical Pharmacists Who Guide Antimicrobial Stewardship
Clinical pharmacists have become an integral part of the medical team in many hospitals, and their impact on clinical and economic outcomes in general has been well documented.14 In regard to antimicrobial stewardship, the Infectious Diseases Society of America (IDSA) and the Society for Healthcare Epidemiology of America (SHEA) guidelines list clinical pharmacists with infectious disease training as core members of the antimicrobial stewardship team (AST).15 In the past, the responsibilities of clinical pharmacists on an AST have focused predominantly on policing, guideline adherence, and formulary management. But recently, IDSA/SHEA guidelines recommended that clinical pharmacists take a more hands-on approach by actively monitoring patients and streamlining or de-escalating empiric antibiotic therapy as soon as culture and sensitivity results are reported.15 One study noted in the guidelines evaluated the impact of an AST with a clinical pharmacist and found that of 625 patients receiving combination antimicrobial therapy, streamlining recommendations were made in 54% of antimicrobial courses, leading to a cost savings of over $100,000 per year.16
The implementation of rapid molecular and microbiological tests will allow clinical pharmacists to recommend streamlining antimicrobial therapy or changing inappropriate therapy sooner, hence improving its value. This also leads to improved clinical outcomes by assuring that patients are on appropriate therapy sooner and helps minimize the risk of adverse reactions and antimicrobial resistance by reducing patients’ exposure to unnecessary antimicrobials. It also allows clinical pharmacists to recommend more cost-effective treatment options (i.e., fluconazole vs. an echinocandin for C albicans infections).
While rapid molecular and microbiological tests provide faster results, their value is undermined when the results and interventions are not made in a timely fashion. A study done by Carver et al found that the use of mecA PCR testing to rapidly identify MRSA in BSIs was being underutilized by physicians, and that using a clinical pharmacist to alert physicians and make clinical recommendations resulted in a 25.4 hour reduction in time to optimal antimicrobial therapy (64.7 ± 36.8 vs. 39.3 ± 15.5 h; P = .002).17 This reduction in time to receipt of optimal antimicrobial therapy could lead to improved clinical outcomes and decreased LOS. Lodise et al found that delayed therapy occurring after 44.75 hours for patients being treated for hospital-acquired S aureus bacteremia resulted in a 3.8 times higher incidence of infection-related mortality and increased length of hospitalization from 14.3 days with timely therapy versus 20.2 days with delayed therapy.18
Conclusion
Despite the lack of published studies evaluating the impact of rapid molecular and microbiological techniques on clinical outcomes and cost avoidance when used for BSIs, the need to identify pathogens and susceptibility faster than standard culture-based techniques is driving their introduction into the clinical setting. PNA FISH is currently the only rapid molecular or microbiological technique with published studies describing clinical and economic benefits, but studies evaluating other types of technology, such as Tem-PCR, that provide clinicians with more detailed information are currently under way. As more rapid molecular and microbiological techniques come to market, it will be important for physicians, clinical pharmacists, and the microbiology community to work collaboratively to fully evaluate the true utility of these tests in the clinical setting and the impact they will have on health care practices going forward.
REFERENCES
1. Kollef MH, Napolitano LM, Solomkin JS. Health care associated infection (HAI): a critical appraisal of the emerging threat—proceedings of the HAI summit. Clin Infect Dis. 2008;47(suppl 2):S55-S99.
2. Minino AM, Smith BL. Deaths: preliminary data for 2000. Natl Vital Stat Rep. 2001;49:1-40.
3. Diekema DJ, Beekmann SE, Chapin KC, et al. Epidemiology and outcome of nosocomial and community-onset bloodstream infection. J Clin Microbiol. 2003;41:3655-3660.
4. Shorr AF, Tabak YP, Killian AD, et al. Healthcare-associated bloodstream infection: a distinct entity? Insights from a large U.S. database. Crit Care Med. 2006;34:2588-2595.
5. Khatib R, Saeed S, Sharma M, et al. Impact of initial antibiotic choice and delayed appropriate treatment on the outcome of Staphylococcus aureus bacteremia. Eur J Clin Microbiol Infect Dis.
2006;25:181-185.
6. Espy MJ, Uhl JR, Sloan LM, et al. Real-time PCR in clinical microbiology: applications for routine laboratory testing. Clin Microbiol Rev. 2006;19:165-256.
7. Lehmann LE, Hunfeld KP, Emrich T, et al. A multiplex real-time PCR assay for rapid detection and differentiation of 25 bacterial and fungal pathogens from whole blood samples. Med Microbiol Immunol. 2008;197:313-324.
8. Tang Y, Kilic A, Yang Q, et al. StaphPlex system for rapid and simultaneous identification of antibiotic resistance determinants and Panton-Valentine leukocidin detection of Staphylococci from positive blood cultures. J Clin Microbiol. 2007;45:1867-1873.
9. Procop GW. Molecular diagnostics for the detection and characterization of microbial pathogens. Clin Infect Dis. 2007;45(suppl 2):S99-S111.
10. Forrest GN, Mehta S, Weekes E, et al. Impact of in situ hybridization on coagulase-negative staphylococci positive blood cultures. J Antimicrob Chemother. 2006;58:154-158.
11. Forrest GN, Mankes K, Jabra-Rizk MA, et al. Peptide nucleic acid fluorescence in situ hybridization-based identification of Candida albicans and its impact on mortality and antifungal therapy costs. J Clin Microbiol. 2006;44:3381-3383.
12. Ly T, Gulia J, Pyrgos V. Impact upon clinical outcomes of translation of PNA FISH-generated laboratory data from the clinical microbiology bench to bedside in real time. Ther Clin Risk Manag. 2008;4:637-640.
13. Forrest GN, Roghmann MC, Toombs LS, et al. Peptide nucleic acid fluorescent in situ hybridization for hospital-acquired enterococcal bacteremia: delivering earlier effective antimicrobial therapy. Antimicrob Agents Chemother. 2008;52:3558-3563.
14. De Rijdt, Willems L, Simoens S. Economic effects of clinical pharmacy interventions: a literature review. Am J Health Syst Pharm. 2008;65:1161-1172.
15. Tonna AP, Stewart D, West B, et al. Antimicrobial optimization in secondary care: the pharmacist as part of a multidisciplinary antimicrobial programme—a literature review. Int J Antimicrob Agents. 2008;31:511-517.
16. Dellit TH, Owens RC, McGowan JE Jr, et al. Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America guidelines for developing an institutional program to enhance antimicrobial stewardship. Clin Infect Dis. 2007;44:159-177.
17. Carver PL, Lin S, DePestel DD, Newton DW. Impact of mecA gene testing and intervention by infectious disease clinical pharmacists on time to optimal antimicrobial therapy for Staphylococcus aureus bacteremia at a university hospital. J Clin Microbiol. 2008;46:2381-2383.
18. Lodise TP, McKinnon PS, Swiderski L, Rybak, MJ. Outcomes analysis of delayed antibiotic treatment for hospital-acquired Staphylococcus aureus bacteremia. Clin Infect Dis. 2003;36:1418-1423.
19. Klouche M, Schroder U. Rapid methods for diagnosis of bloodstream infections. Clin Chem Lab Med. 2008;46:888-908.





