ABSTRACT: Nonsteroidal anti-inflammatory drugs (NSAIDs) are commonly used for management of inflammation and pain, but they can cause gastrointestinal, cardiovascular, and renal adverse effects. Genetic variations in CYP450 enzymes affect their metabolic activity, thus impacting the hepatic clearance, elimination half-life, and risk of adverse effects of drugs metabolized by that CYP. Celecoxib, ibuprofen, flurbiprofen, meloxicam, and piroxicam are significantly metabolized by CYP2C9. The Clinical Pharmacogenetic Implementation Consortium has published evidence-based recommendations on CYP2C9 polymorphism–based safe usage of NSAIDs. As pharmacogenetics becomes more common, pharmacists will play a critical role in optimizing drug regimens based on pharmacogenetic test results.
A variety of patient-specific factors, such as age, sex, body weight, diet, disease conditions, and smoking status, can affect an individual’s response to drug therapy. Pharmacogenetics is an emerging field focused on how an individual’s genetic makeup affects the pharmacodynamic response, pharmacokinetics, or risk of adverse reactions to drugs.1,2 In 2003, the Human Genome Project was completed to map and gain a deeper understanding of the human genome. Although the disparities in drug response in various ethnic backgrounds have been known for decades, our understanding of the genetic basis of these variations in drug response has grown exponentially in recent years after the completion of the Human Genome Project.
Despite the vast knowledge of genetic variation’s effect on drug response or risk of side effects, there was a lack of guidance on how to use this information for improving patient care clinically. Therefore, the international Clinical Pharmacogenetics Implementation Consortium (CPIC) was formed in 2009 to provide actionable guidance based on available pharmacogenetic knowledge. CPIC publishes evidence-based, peer-reviewed clinical-practice guidelines for implementing individualized therapy for patient care based on pharmacogenetic tests.3 CPIC guidelines include a systemic grading of prospective clinical evidence and actionable clinical recommendations.3
Clinical pharmacogenetic tests targeted to identify these specific genetic variations are increasingly available. The Genetic Testing Registry (www.ncbi.nlm.nih.gov/gtr/) is a useful resource for clinicians in determining the availability of a clinical pharmacogenetic test for their patients. The resource includes the pharmacogenetic test’s purpose, methodology, validity, evidence of the test’s usefulness, and laboratory contacts. While CPIC provides guidance on implementing an actionable pharmacogenetic result, it does not address who should be tested. The FDA has also begun to include pharmacogenetic information in package inserts. These now offer pharmacogenetic information on over 400 medications, including boxed warnings that recommend pharmacogenetics testing before initiating certain medications, like abacavir. Thus, recent federal and international efforts have been playing a more prominent role in optimizing individualized drug therapy to improve both safety and efficacy by applying pharmacogenetics knowledge.
Genetic Polymorphism
Genetic polymorphism plays an essential role in the process of natural selection and evolution. Polymorphism is the occurrence of two or more variations in the DNA sequences at a given locus of a gene with a frequency greater than 1% in the population.4-6 Such gene variants, alleles, can change the structure of an encoded protein or protein expression, or they may not affect the protein function. Everyone has two copies of each allele, one from the biological mother and one from the biological father. The two identical alleles are classified as homozygous, while two different alleles are classified as heterozygous. While polymorphism is vital for creating genetic diversity, allelic variations occurring at less than 1% frequency in the population and typically contributing to diseases are termed mutations.4-6 Polymorphism arises in the germline cells, meaning it originates from the reproductive cells of biological parents and is present in every nucleated cell in the body. Similar to polymorphism, mutations can also be in the germline cells and are heritable (e.g., hemophilia, sickle cell anemia). However, they can also be in the somatic cells localized to a specific tissue originating during the lifetime and nonheritable.7,8 Many such local tissue-specific mutations are found in cancers. Such mutations form the basis of tumor genomics and make them amenable to the selection of personalized therapies based on pharmacogenetic testing of the tumor genome.
Four major types of DNA variability that cause polymorphism or mutations include 1) single nucleotide polymorphism (SNP), which is a substitution, insertion, or deletion of a single nucleotide; 2) insertion or deletion (indel) of more than one nucleotide ranging from a few to several hundred nucleotides; 3) variation in the number of tandem repeats; and 4) gene duplication. Among these, SNPs are most common allelic variations accounting for genetic polymorphism.9-11 Genetic polymorphism in drug-metabolizing enzymes, drug transporters, and drug targets have all been shown to contribute to variation in drug response.1,2 Significant advancements have been achieved in understanding the polymorphic variation in drug-metabolizing enzymes, such as hepatic cytochrome P450 (CYP) enzymes and phase two drug-metabolizing enzymes like UGT1A1 and TPMT, and the impact on the efficacy and safety of drugs metabolized by these enzymes.
CYP450 Polymorphism Genotypes and Phenotypes
CYP450 enzymes play a major role in drug metabolism. The Human Genome Project has identified 57 human CYPs; however, 90% of drugs are metabolized by six of those enzymes, CYP1A2, CYP2C9, CYP2C19, CYP2D6, CYP3A4, and CYP3A5.12 Over 2,000 allelic variations have been observed in these CYP enzymes, most of which occur due to SNPs.13 Genetic polymorphism in CYP enzymes typically contributes to a decrease or loss of CYP enzyme activity.12-16 Although the field of pharmacogenetics uses many different nomenclatures to report polymorphism, star allele nomenclature is used for CYP polymorphism. Wild-type allele with normal enzymatic activity is the most frequently occurring allele in the population and is always assigned as *1 allele (e.g., CYP2C9*1 or CYP2C19*1).12-16 Other alleles with or without altered enzymatic activity are assigned different star numbers, typically in the order of discovery.12-16
A decrease in any given CYP enzyme activity impacts the hepatic clearance of drugs that are metabolized by that CYP enzyme, thus affecting the plasma concentration and elimination half-life of those drugs. Patients carrying homozygous (two identical copies) wild-type CYP alleles with normal enzyme activity have the normal-metabolizers (also known as extensive) phenotype. However, patients who have heterozygous genotypes with one copy of the wild-type allele and one copy of a reduced-function or loss-of-function allele or who have homozygous with two copies of reduced-function allele would be classified as the intermediate-metabolizers phenotype. Those who are homozygous with two copies of loss-of-function alleles or heterozygous with one copy of loss-of-function alleles and one copy of reduced-function allele would be classified as poor-metabolizers phenotype.
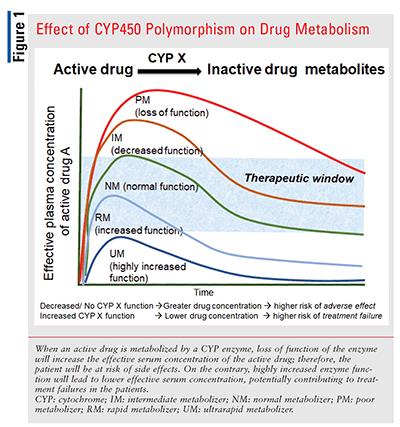
In patients who are CYP intermediate or poor metabolizers, a drug metabolized by that CYP enzyme will not be metabolized as quickly as normal metabolizers.12,15 Thus, the effective drug concentration will remain high, and the risk of an adverse effect will be greater for intermediate or poor metabolizers (FIGURE 1). Any prodrug that is converted to its active metabolite by a CYP enzyme may not be converted to effective therapeutic concentrations by intermediate or poor metabolizers, and there will be a risk of therapeutic failure. One such example is clopidogrel that is converted to its active metabolite by CYP2C19. Patients carrying loss-of-function CYP2C19 alleles are at increased risk of thrombotic events despite taking the label-recommended dose.
Rarely, allelic variation in CYP can cause an increase in function (e.g., CYP2C19*17 allele). Patients with heterozygous or homozygous genotypes for increased-function CYP alleles have rapid- and ultrarapid-metabolizer phenotypes and will metabolize the drug faster. In these cases, the effective plasma therapeutic concentration may not be achieved, resulting in a risk of treatment failure for the rapid and ultrarapid metabolizers.
Nonsteroidal Anti-Inflammatory Drugs
Nonsteroidal anti-inflammatory drugs (NSAIDs) act by inhibiting cyclooxygenase (COX), the enzyme responsible for the formation of proinflammatory autacoids called prostaglandins by catalyzing arachidonic acid.16 Nonselective NSAIDs inhibit both COX-1 and COX-2, whereas selective NSAIDs are exclusively COX-2 inhibitors.17 NSAIDs are commonly used as antipyretic, analgesic, and anti-inflammatory agents worldwide. NSAIDs are the first-line agents for mild-to-moderate musculoskeletal pain and inflammation for short-term use, as well as chronic noncancer nociceptor pain in conditions such as rheumatoid arthritis for long-term use. There are currently more than 20 oral NSAIDs available in the United States; ibuprofen, meloxicam, and celecoxib are among the top 200 most commonly prescribed.
At equipotent doses, the responses to NSAIDs and their associated risk of adverse effects show interpatient variability owing to pharmacodynamic and pharmacokinetic factors, including pharmacogenetics. Although NSAIDs offer many advantages over opioids and steroids, such as the absence of addictive potential and lower incidence of adverse effects, serious side effects can still be attributed to them, especially when significant drug-drug interactions or relevant comorbidities are present.18-20
Gastrointestinal complications are the most common side effect of nonselective NSAIDs because of the inhibition of COX-1 in the gastric mucosa due to the reduced formation of gastric prostacyclins.18-20 Studies have shown that gastrointestinal bleeding and ulcers develop in 1% to 2% and 15% to 30% of patients, respectively, who regularly take nonselective NSAIDs.18-20 Although the selective NSAIDs, such as celecoxib, meloxicam, and diclofenac, have a lower risk of gastrointestinal adverse effects, they have a greater risk of cardiovascular events due to a likely imbalance between antithrombotic and prothrombotic prostaglandins such as thromboxane and prostacyclin.18-20 In addition to gastrointestinal bleeding, myocardial infarction, and sudden cardiac arrest, chronic use of NSAIDs may also lead to hypertension, heart failure, renal damage, and arrhythmias.18-20
Given the large consumption of NSAIDs, these side effects have considerable health and economic impacts. These adverse effects can be seen to a greater extent in older patients and patients with pre-existing gastrointestinal disease, cardiovascular disease, chronic kidney disease, chronic liver disease, or aspirin-exacerbated respiratory disease.18-20 Besides these risk factors, recent evidence has shown that CYP polymorphism can also significantly impact NSAID exposure and thus amplify the risk of side effects.21 Therefore, the clinical pearl of prescribing NSAIDs at the “lowest effective dose for the shortest duration of time” may not be safe for some patients, and pharmacogenetics knowledge can help identify these patients.
Impact of CYP2C9 Polymorphism on Oral NSAID Metabolism
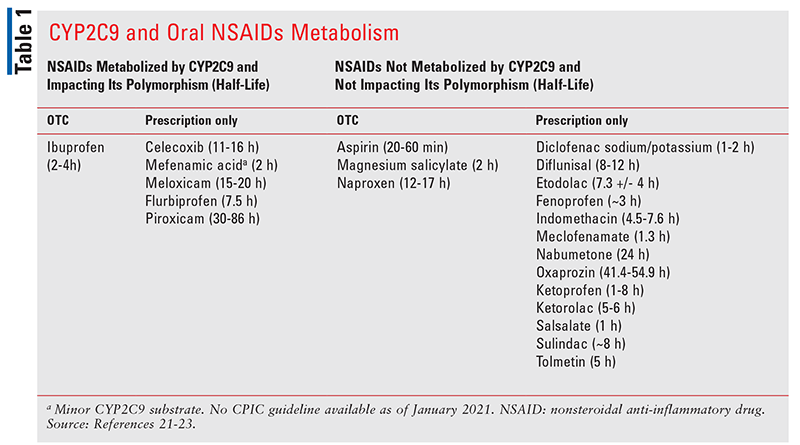
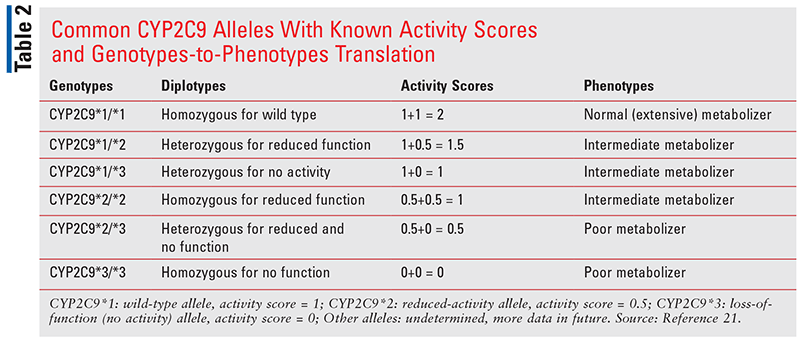
CYP2C9-mediated hepatic metabolism plays a significant role in the inactivation of specific NSAIDs, such as celecoxib, meloxicam, ibuprofen, flurbiprofen, and piroxicam (TABLE 1).21-23 CYP2C9 is highly polymorphic, with a total of 61 alleles reported, all of which have either reduced or no enzymatic activity.21 A patient’s phenotype is calculated based on the genotype of the CYP2C9 allele, its heterozygosity or homozygosity (also referred to as diplotype). Based on the allelic function, an activity score representing the function is assigned.21
For example, the wild-type CYP2C9*1 allele has been assigned an activity score of 1, CYP2C9 alleles (e.g., CYP2C9*2) that cause a decrease in enzyme function are assigned an activity score of 0.5, and alleles (e.g., CYP2C9*3) with complete loss of enzyme activity have a score of 0. The activity scores of both alleles present in the patient are added together to determine the activity score of the diplotype, and a phenotype is determined from the resulting diplotype activity score (TABLE 2).21 For example, a patient carrying both wild-type alleles will have an activity score of 2 and a normal-metabolizer phenotype. Patients with one copy of wild-type and one copy of reduced-function allele will have an activity score of 1.5, and patients with two copies of reduced-function or one copy of wild-type and one copy of loss-of-function allele will have an activity score of 1. Both activity scores, 1.5 and 1, are considered intermediate-metabolizer phenotype.
Lastly, a patient carrying one copy of the reduced-function allele and one copy of loss-of-function allele will have an activity score of 0.5, and if two copies of loss-of-function allele are present, then an activity score of 0. Both 0.5 and 0 activity scores are assigned as poor-metabolizer phenotype (TABLE 2). 21
Clinical Recommendations for Using NSAIDs in CYP2C9 Polymorphism
Recently, CPIC conducted a systematic review of the published literature investigating the effect of CYP2C9 polymorphism on the plasma AUC of various NSAIDs and published guidelines on dose adjustment or selection of alternative NSAIDs in patients carrying reduced-function or loss-of-function CYP2C9 alleles.21
The half-life of a drug, an essential determinant of the AUC, and any factor that decreases the drug metabolism will have a significant impact on increasing the AUC of the drug with a longer half-life. Therefore, CYP2C9 polymorphism has a significantly higher impact on increasing the AUC of NSAIDs with longer half-lives, namely meloxicam and piroxicam, compared with the NSAIDs with a shorter half-life, such as celecoxib, ibuprofen, and flurbiprofen.
A meta-analysis of clinical studies showed that as compared with individuals carrying wild-type CYP2C9*1/*1 (normal metabolizers; activity score 2), ratios of AUC mean of ibuprofen (half-life 2-4 hours), celecoxib (half-life 11-16 hours), and meloxicam (half-life 15-20 hours) in patients with CYP2C9 *1/*3 (intermediate metabolizers; activity score 1) increased to 1.43, 1.62, and 1.82, respectively.21 For CYP2C9*3/*3 carriers (poor metabolizers; activity score 0), the ratio of AUC means was significantly increased, 4.17, in patients treated with celecoxib.2 Therefore, CPIC recommendations for NSAIDs based on CYP2C9 polymorphism vary depending upon the half-life of NSAIDs.21
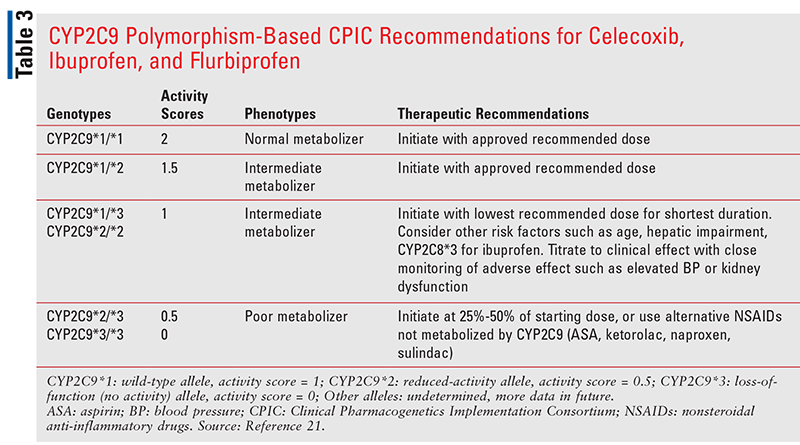
For the NSAIDs with short-to-moderately-long half-lives (ibuprofen 2-4 hours, flurbiprofen 2-6 hours, and celecoxib 11-16 hours), the CPIC guidelines recommend that patients with normal-metabolizers phenotype (activity score 2) and intermediate-metabolizers phenotype with an activity score of 1.5 can initiate treatment at the label-recommended dose (TABLE 3).21 In intermediate-metabolizers phenotype with an activity score of 1, treatment should be initiated with the lowest recommended dose and for the shortest duration.21 Other risk factors, such as age and hepatic impairment, must also be considered, and these patients should also be closely monitored for adverse effects. For poor metabolizers (activity score 0.5 or 0), ibuprofen, flurbiprofen, and celecoxib should be initiated at 25% to 50% of the starting dose (TABLE 3),21 or alternative NSAIDs that are not metabolized by CYP2C9 should be used in those patients (TABLE 1).21
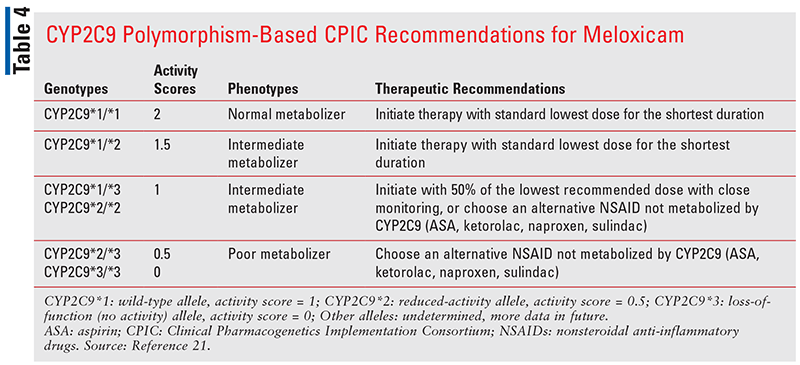
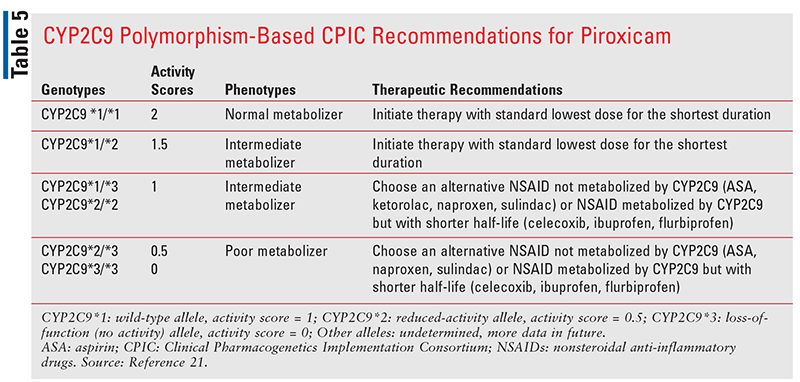
For meloxicam (half-life 15 to 20 hours) and piroxicam (half-life 30 to 86 hours), the CPIC guideline recommendation for normal metabolizers and intermediate metabolizers with an activity score of 1.5 is to initiate treatment with the lowest dose for the shortest duration (TABLES 4 ,5).21 For intermediate metabolizers with an activity score of 1, meloxicam can be initiated with 50% of the lowest recommended dose with close monitoring, or an alternative NSAID that is not metabolized by CYP2C9 should be used.21
For piroxicam, which has a half-life even longer than meloxicam, it is recommended that CYP2C9 intermediate metabolizers with an activity score of 1 should not be given piroxicam (TABLE 5).21 An alternative NSAID that is not metabolized by CYP2C9 (TABLE 1) or an NSAID metabolized by CYP2C9 with a shorter half-life should be used in such patients. Patients with CYP2C9 poor-metabolizer phenotype (activity score 0.5 or 0) should not receive either meloxicam or piroxicam. Instead, an alternative NSAID that is either not metabolized by CYP2C9 or an NSAID metabolized by CYP2C9 with a shorter half-life should be considered.21
Lastly, NSAIDs may be used on a chronic, short-term, or as-needed basis. The CPIC recommends that these guidelines be applied regardless of treatment duration, and also for OTC NSAIDs, since the risks of upper gastrointestinal bleeding and myocardial infarction rates are thought to be similar among new and chronic NSAID users.21,24,25 NSAIDs are also commonly used in pediatric patients to reduce fever and alleviate pain and in premature infants or neonates with patent ductus arteriosus, to close the ductus. The CYP2C9 enzyme activity is fully mature by early childhood. Therefore, it may be appropriate to use these recommendations for adolescents and younger children, along with close monitoring.21 Overall, more research is needed in the pediatric population on the relationship between CYP2C9 genotypes and NSAID treatment outcomes.
Role of Pharmacists in CYP2C9-Guided NSAID Dosing
Although NSAIDs are generally considered safe, there is known significant interpatient variability due to CYP2C9 polymorphism. The CYP2C9 polymorphism status of a patient is most beneficial for clinical actionability at the start of new NSAIDs. Despite the actionability of CYP2C9 polymorphism in NSAID dosing, routine pharmacogenetic testing of CYP2C9 is rarely done in clinical practice prior to NSAID start. The FDA currently does not provide any guidance on testing CYP2C9 for any NSAIDs in their package labels. However, with the increasing implementation of pharmacogenetics, the application of known CYP2C9 altered-metabolizer statuses will become more common. A more likely scenario of using CYP2C9 polymorphism in NSAID dosing is when patients already have had pharmacogenetic testing done for other clinical reasons.
Germline polymorphism such as CYP2C9 polymorphism remains unchanged throughout the patient’s lifetime, and pharmacogenetic testing results can be reused. Pharmacists will often be responsible for reevaluating patients with available pharmacogenetic testing results when NSAIDs are prescribed or being considered. Since pharmacogenetic knowledge is growing rapidly, we recommend pharmacists use the latest CPIC guidelines for the most up-to-date recommendation. PharmGKB (www.pharmgkb.org) is an excellent Web-based resource that regularly updates CYP polymorphism information, activity-score calculation, and clinical guidelines from various initiatives and can be used as the most up-to-date pharmacogenetics clinical resource.26
Conclusion
NSAIDs are among the most commonly used drugs, with a variety of approved indications in the U.S. Pharmacogenetic knowledge of CYP2C9 polymorphism and other factors, such as half-life, may help clinicians to identify patients at risk for serious adverse effects and avoid NSAIDs, as necessary. As pharmacogenetic knowledge continues to grow and clinicians start to adopt pharmacogenetic recommendations in routine clinical practice, pharmacists will play an active role in implementing pharmacogenetic guidelines. It is recommended that pharmacists use the most up-to-date resources for implementation.
The content contained in this article is for informational purposes only. The content is not intended to be a substitute for professional advice. Reliance on any information provided in this article is solely at your own risk.
REFERENCES
1. Eichelbaum M, Ingelman-Sundberg M, Evans WE. Pharmacogenomics and individualized drug therapy. Annu Rev Med. 2006;57:119-137.
2. Weinshilboum RM, Wang L. Pharmacogenomics: precision medicine and drug response. Mayo Clin Proc. 2017;92:1711-1722.
3. Caudle KE, Klein TE, Hoffman JM, et al. Incorporation of pharmacogenomics into routine clinical practice: the Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline development process. Curr Drug Metab. 2014;15:209-217.
4. Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet. 2006;7:85-97.
5. Wright AF. Genetic variation: polymorphisms and mutations. In: Nature Encyclopedia of the Human Genome. London: Nature Publishing Group; 2003:959-968.
6. Karki R, Pandya D, Elston RC, Ferlini C. Defining “mutation” and “polymorphism” in the era of personal genomics. BMC Med Genomics. 2015;8:37.
7. Griffiths AJF, Miller JH, Suzuki DT, et al. An Introduction to Genetic Analysis. 7th edition. New York: W. H. Freeman; 2000.
8. Milholland B, Dong X, Zhang L, et al. Differences between germline and somatic mutation rates in humans and mice. Nat Commun. 2017;8:15183.
9. Krynetskiy E. Beyond SNPs and CNV: Pharmacogenomics of polymorphic tandem repeats. J Pharmacogenomics Pharmacoproteomics. 2017;8:170.
10. O’Dushlaine CT, Edwards RJ, Park SD, Shields DC. Tandem repeat copy-number variation in protein-coding regions of human genes. Genome Biol. 2005;6:R69.
11. Schrider DR, Hahn MW. Gene copy-number polymorphism in nature. Proc Biol Sci. 2010;277:3213-3221.
12. Lynch T, Price A. The effect of cytochrome P450 metabolism on drug response, interactions, and adverse effects. Am Fam Physician. 2007;76:391-396.
13. Preissner SC, Hoffmann MF, Preissner R, et al. Polymorphic cytochrome P450 enzymes (CYPs) and their role in personalized therapy. PLoS One. 2013;8:e82562.
14. Tomalik-Scharte D, Lazar A, Fuhr U, Kirchheiner J. The clinical role of genetic polymorphisms in drug-metabolizing enzymes. Pharmacogenomics J. 2008;8:4-15.
15. Tornio A, Backman JT. Cytochrome P450 in pharmacogenetics: an update. Adv Pharmacol. 2018;83:3-32.
16. Ingelman-Sundberg M. Pharmacogenetics of cytochrome P450 and its applications in drug therapy: the past, present and future. Trends Pharmacol Sci. 2004;25:193-200.
17. Grosser T, Smyth E, FitzGerald G. Pharmacotherapy of inflammation, fever, pain, and gout. In: Brunton, LL, Hilal-Dandan R, Knollmann BC, eds. Goodman & Gilman’s: The Pharmacological Basis of Therapeutics, 13th ed. New York, NY: McGraw-Hill; 2018:685-710.
18. Wongrakpanich S, Wongrakpanich A, Melhado K, Rangaswami J. A comprehensive review of nonsteroidal anti-inflammatory drug use in the elderly. Aging Dis. 2018;9:143-150.
19. Ong CK, Lirk P, Tan CH, Seymour RA. An evidence-based update on nonsteroidal anti-inflammatory drugs. Clin Med Res. 2007;5:19-34.
20. Davis A, Robson J. The dangers of NSAIDs: look both ways. Br J Gen Pract. 2016;66:172-173.
21. Theken KN, Lee CR, Gong L, et al. Clinical Pharmacogenetics Implementation Consortium guideline (CPIC) for CYP2C9 and nonsteroidal anti-inflammatory drugs. Clin Pharmacol Ther. 2020;108:191-200.
22. Lexi-Drugs. Lexicomp. Riverwoods, IL: Wolters Kluwer Health, Inc. http://online.lexi.com. Accessed January 10, 2020.
23. Micromedex Solutions. Ann Arbor, MI: Truven Health Analytics, Inc. http://www.micromedexsolutions.com. Accessed January 10, 2020.
24. Schjerning Olsen AM, Fosbøl EL, Lindhardsen J, et al. Duration of treatment with nonsteroidal anti-inflammatory drugs and impact on risk of death and recurrent myocardial infarction in patients with prior myocardial infarction: a nationwide cohort study. Circulation. 2011;123:2226-2235.
25. Hernandez-Diaz S, Garcia-Rodriguez LA. Epidemiologic assessment of the safety of conventional nonsteroidal anti-inflammatory drugs. Am J Med. 2001;110(suppl. 3A): 20S-S27.
26. Whirl-Carrillo M, McDonagh EM, Hebert JM, et al. Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther. 2012;92:414-417.
To comment on this article, contact rdavidson@uspharmacist.com.